A new visualization tool helps people understand the ways that full-siblings can share DNA as well as average sharing percentages
Yesterday a new tool was released that will help a lot of beginners and veteran genetic genealogists alike. These visualizations show in the simplest way possible why the full-sibling averages of shared DNA are 50%, 37.5%, or 25% depending on which metric is used. It will also make it much easier to understand DNA basics such as the fact that people usually have two copies of autosomal chromosomes, one paternal and one maternal.
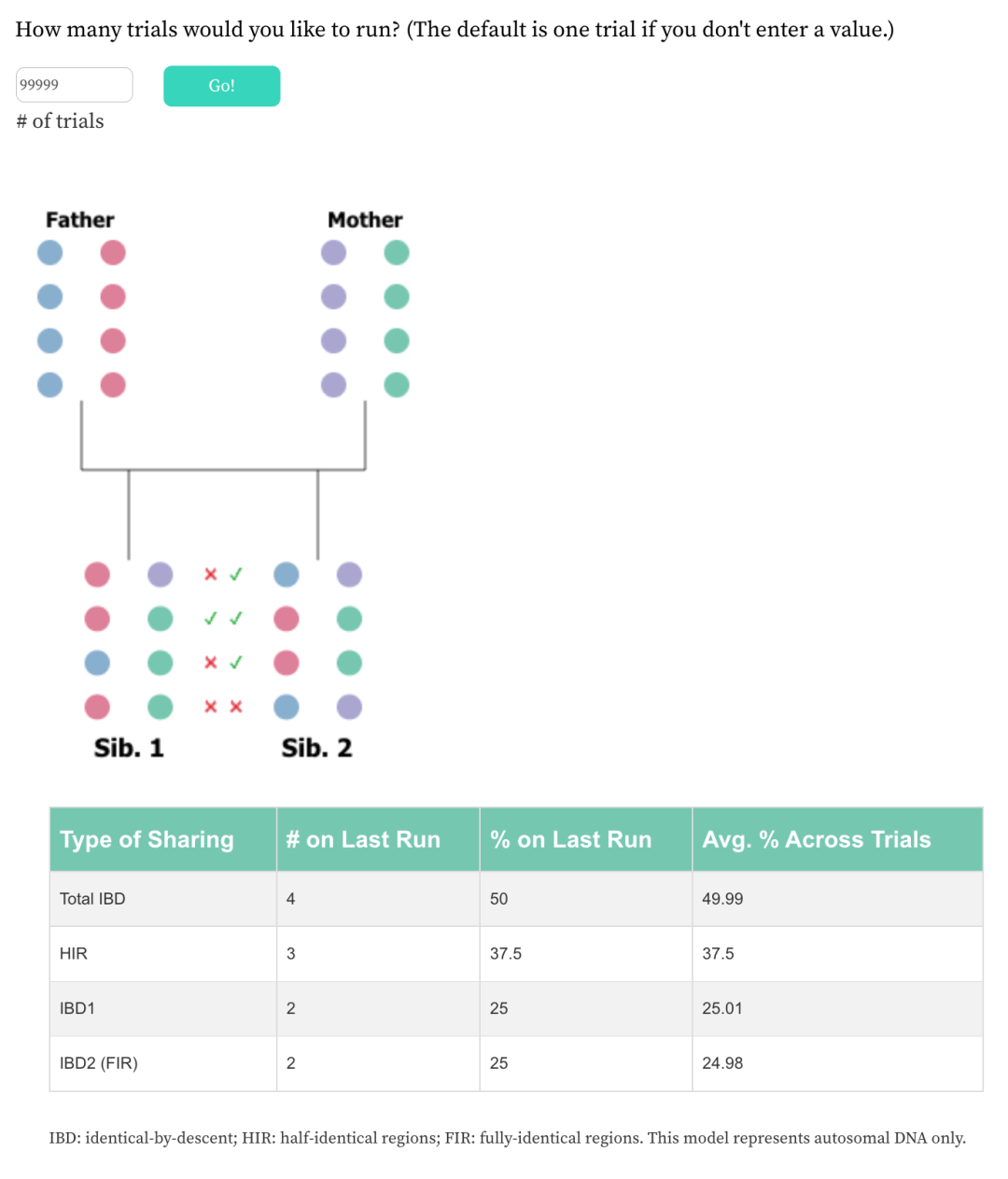
Sibling Lotto lets you do one trial run or up to 1M trials at once. No matter how many trials you choose, the results from the last one will be displayed. But you can also see aggregate statistics from the rest of the trials.
Here is a graphic from the Sibling Lotto showing a trial run that happened to have an average result.  The average shared DNA between full-siblings is something that isn’t talked about much. When there is a conversation about it, it usually centers on the confusion of whether or not the full-sibling average is 37.5% or 50%. These conversations used to go on without making anything clearer until an article came out in 2021 that a lot of people have found helpful. Key to the issue is the fact that different metrics are used by different DNA sites.
The average shared DNA between full-siblings is something that isn’t talked about much. When there is a conversation about it, it usually centers on the confusion of whether or not the full-sibling average is 37.5% or 50%. These conversations used to go on without making anything clearer until an article came out in 2021 that a lot of people have found helpful. Key to the issue is the fact that different metrics are used by different DNA sites.
You can see in the graphic that one quarter of the siblings’ circles come from rows that have fully-identical DNA. There won’t always be such a row in this very simple model, but it happened in the second row this time. If we count those two checkmarks and then divide that by eight, which is the total number of circles, we see that 25% of the DNA is from fully-identical regions (FIR), also called IBD2. This is the average amount DNA and full-siblings should always share at least some FIR DNA.
We can also get the amount of total IBD sharing from the above graphic. This can be obtained by adding up all of the checkmarks in both columns and then dividing again by eight. This time it’s 4/8, or 50%. This is again the average for this metric. It’s important to note that total IBD sharing doesn’t have an average of 37.5%. That value can be found in a different metric.
Half-identical region sharing (HIR) is how centiMorgans are reported at most DNA sites (all except 23andMe). You can obtain the HIR value in the above graphic by counting the number of rows that contain at least one checkmark and then dividing by eight. In this case it’s 3/8ths, or 37.5%, which is the true average of HIR sharing for full-siblings. Please note that AncestryDNA has been reporting percentages as a range of total IBD (e.g. 47-53%) for the past couple of years, while still reporting cMs as HIR values.
There’s only one metric left: IBD1 sharing. You can obtain that value by counting the number of rows that have only one checkmark, so this time you exclude the checkmarks in FIR rows. For IBD1 sharing, the graphic above contains two rows. Two divided by eight gives us 25%, which is also the average amount of IBD1 sharing for full-siblings in real life. The IBD1 metric is useful along with the FIR amount for calculating the other metrics. The total IBD value can be obtained by adding IBD1 and FIRs. And the HIR value can be obtained by adding IBD1 and half of the FIR DNA.
You don’t have to calculate all of the above values the way that I’ve done above. Sibling Lotto will actually do all of that for you. Below is another screenshot from the tool. This time the last trial run once again shows average values. And those percentages are reflected in a table below the graphic. But we also see the averages from 99,999 trials in the column to the right. It’s easy to see that the percentages are converging towards the known, theoretical averages.
I find that it usually takes about four clicks of the button before I arrive at a graphic that shows the averages for all metrics of shared DNA between full-siblings, but it can often happen in one click or many more than four. How many times do you have to click the button before you get average results? You can leave a comment here with your answer.
The inherited DNA from parents shown here is random. There are only a few rules for this model. One is that a circle in a given sibling row has to come from the same parent row, i.e. a sibling gets their row one circle from row one of a parent. Another is that a sibling’s left column comes from their father and their right column comes from their mother. The only other rule is that each circle in a sibling’s rows and columns has an equal chance of coming from the applicable parent’s maternal or paternal column, i.e. a circle in a sibling’s first first column has a 50% chance of being from their father’s paternal column and a 50% of coming from their father’s maternal column.
Since this model is simplified to the fewest number of circles (four per column) possible to allow for the possibility of a trial run with the same average values of shared DNA seen in real sibling pairs, we will see extreme examples far more often than in real life. For example, we will often see sibling pairs who share no FIR or 50% or more FIR. And we’ll sometimes see sibling pairs who share no DNA or who share at least half-identical DNA over the whole length of the genome. Conclusions can’t be drawn from the variability shown in the table; only from the averages if enough trials were done. The variability is high because of the low number of circles that were used (four) per column of the genome. Using eight circles would decrease the variability and still allow for trial runs with the exact average. Using many more circles would decrease the variability to be even lower than what it is in real life. (The standard deviation for shared DNA between full-siblings is about 3.6%.)
When using Sibling Lotto, we can now see why fully-identical regions need to be counted twice rather than once in order to calculate total IBD sharing: When there are matching circles on both the left and the right, you wouldn’t count just the left side (paternal) circles as a match but not the right side (maternal) circles. Another way to say it is that, for rows with two checkmarks, you would want to add both checkmarks to get total IBD sharing. Sometimes centiMorgans are reported as HIR values, which are the IBD1 values plus only one copy of the IBD2 DNA. It’s sometimes fine to do it that way, but it’s important to remember that that isn’t the total amount of IBD sharing.
Sibling Lotto is designed to help people understand the types of IBD sharing in the simplest way possible. Developing an understanding of this tool will show that a concept that most people find complicated is actually very simple. I’ve seen a lot of questions from people who were trying to understand the averages of IBD sharing between full-siblings. Even some influential people in the genetic genealogy community have asked me multiple times to explain how to obtain these averages. At those times I always wanted to be able to point to graphics like those above for an average case, but none were available.
This is probably the silliest genetic genealogy tool I’ve made, so I have no doubt it will be the most popular. If you’re interested in more serious work, check out the most accurate and advanced relationship predictions available:
I hope you enjoy Sibling Lotto! Feel free to share it with those looking for a learning aid about DNA randomness and averages of shared DNA. Tool released 10 Jul. 2022.
DNA-Sci — advancing the science of relationship predictions. Please also submit data to this new DNA match survey that will greatly help improve and build new relationship prediction tools. You can also find mobile apps. for relationship predictions in the Apple Store and on Google Play. Feel free to ask a question or leave a comment. You might also like this tool to visualize how much DNA full-siblings share. DNA-Sci is also the original home of DNA coverage calculations.

Recent Comments