People have been asking for this for a long time. Relationship predictions that include # of segments work much better than expected.
Today a new relationship predictor is available for you to use for free. You can now enter the total number of segments along with total centiMorgans (cMs) to see how you’re related to your DNA matches. I always thought that this would be useful, but it’s a thousand times better than I expected.
Back in November I thought I would give it a try to see what would happen. With the first value I tried, my Python algorithm returned a prediction that I knew was amazing. I’ve since worked tirelessly to improve the predictions further. It’s been very difficult to keep it a secret. Actually, I did help several people along the way without mentioning that a new tool was almost ready.
The new predictor often tells you the exact relationship and gender path for the match you’re viewing, i.e. paternal half-sibling, maternal grandparent/grandchild, or paternal grandparent/grandchild. I’ve now seen countless cases when a person knew that a relationship was, for example, paternal. Then the predictions based on cMs and number of segments end up leaving only one option, such as paternal aunt/uncle.
Updates: Check out this new article with even further validation and some tips for understanding probabilities and what makes a good relationship prediction. And use this tool for even better predictions when siblings have tested.
Empirical data
For validation, I used a dataset of 125 known close family to 1st cousin matches. I’ve asked an algorithm in Python to return six random examples from that dataset. I only did this once. Below I’ll show you the predictor output from the one and only time I’ve taken a random sample of the dataset, in the order that the algorithm gave them to me. I’ve denoted the known relationships in red below each prediction output.






In all but one case only three relationship subtypes are listed as possibilities. This cuts down the possible relationships by half from six. (Actually, predictions for 1,986 cMs would’ve also returned full-siblings as an option if we didn’t enter the number of segments, so there’s another benefit of the new tool.) Reducing the number of possible relationships by half drastically reduces the amount of time that people have to spend finding out the true relationship.
But what’s even better is that the correct relationship subtype is usually the very first one listed because of the very high probabilities. And in the one case in which it wasn’t at the top, maternal half-sibling actually had the highest individual probability! In half of the cases, if you knew the relationship was paternal, then you’d have less than 1% chance that the true relationship was anything other than the first one listed. I saw this happen dozens of times in beta tested data and when people were asking for help online. Ancestry tells most of us which side the match is on now, at least once we’ve designated which side is Parent 1 and which is Parent 2, so that makes it quite easy to know the relationship in a lot of cases.
Of course, people still have to do their due diligence to verify the correct match. This should be done genealogically, where possible. And you should still sort your matches into four groups and verify the right matching patterns. For example, if you think your match is a half-sibling, both of you should match through only one grandparent pair. If your match shows as “both sides,” that suggests that you might be the person’s aunt or uncle. But then sometimes there are double connections—all of us have them if you go back far enough. This complicates sorting our matches. When it’s hard to get your matches into four neat groups, what a boon it is to have a predictor that returns such high probabilities for the correct match!
Speaking of that, the average probability assigned to the correct match across the entire dataset of 125 matching pairs was a whopping 45%! It’s a bit higher, actually. In our random sample of six above we have four that are slightly under that and two that are astoundingly good. That’s well in line with my observations of the dataset on the whole. I’ve seen at least six cases when the probability of the correct relationship had 96% probability or more, including some with 100%.
What kind of probabilities should we expect to get for the correct relationship? When six relationship types are possible for Group 2 (the 25% average, or half-sibling, avuncular, and grandparent/grandchild group), evenly splitting them would give us 100%/6, or about 16.7%. So anything higher than that is good. For Group 3 (the 12.5% average, or 1st cousin group), where 12 different relationship subtypes are possible, predicting the correct relationship at 8% or higher would be good. But then there are cM and segment values for which both Group 2 and Group 3 are possible. We would expect some pretty low probabilities assigned to the correct relationship subtype in these ranges.
And that’s what makes the new tool so amazing, assigning over 45% probability to the correct subtype. Anyone can check their known values against these predictions and I highly encourage that. The vast majority of people who check a few known close family members will see very high probabilities assigned to the correct relationship.
I’d like to show a few more predictions that weren’t part of the random sample above so you can see what else this predictor can do.



The paternal grandfather match is obviously as good as it gets. Imagine being the person who found an unknown match and plugged this amount of cMs and segments into the tool. The tool handles 1st cousin relationships quite well, too, although the more distant the relationship is, the less differences we’ll see in the probabilities. The next match was a known maternal maternal 1st cousin and that relationship is right at the top of the probability list. The third match is a known maternal paternal 1st cousin. Keep in mind that a maternal paternal 1st cousin is the same relationship as a paternal maternal 1st cousin if viewed from the other’s perspective, and so the combined probability for those two would be 30.2%.
All full-sibling and parent/child matches in the empirical dataset were assigned a probability of 100% by the tool. If entering only cMs in two different relationship predictors, two of those full-sibling pairs were given less than 100% probability. Predictions are simply better when including the number of segments.
What led us here?
Background
Really, scientists from 23andMe showed us that the number of segments is important back in 2012. Their scatterplot shows pretty distinct values when comparing different relationships.

I had to wonder if we could get good predictions if we further separated relationships by maternal and paternal sides.
Edit 19 Apr. 2023: Kitty Cooper showed back in 2017 that the number of segments can be useful for telling the difference between relationships and even for paternal/maternal sides.
The figure below shows cMs vs. segments for parent/child, full-sibling, and sex-specific plots for half-sibling, grandparent/grandchild, and aunt/uncle/niece/nephew. It also shows the difference between paternal paternal 1st cousins and maternal maternal 1st cousins.

There’s a lot of overlap between relationship subtypes in the scatterplot, far more than in the 23andMe plot and one has to wonder how theirs got to be so optimistic. Part of the answer might be because the 23andMe plot included far less data. It also appears like the 23andMe plot might only include full-siblings and not half-siblings. The new plot makes it seem as thought it would be hard to give a high probability to any one relationship in Group 2 (25% average group). Yet we often see 50% and sometimes see 100% given to one relationship subtype.
I found it difficult to choose colors such that the entire center of the plot wouldn’t look like one brown blob. Adding transparency for some, I can now see six different clusters. Those relationships, from the top, are red for maternal half-sibling, blue for maternal avuncular (aunt/uncle/niece/nephew), pink for paternal avuncular, purple for paternal half-sibling, grey for maternal grandparent/grandchild, and a different shade of blue for paternal grandparent-grandchild. (Paternal half-sibling markers have less transparency in the plot, which can unrealistically make their cM values look higher than grandparent/grandchild.)
As you can see the paternal relationships have fewer segments than the maternal relationships. This is expected because the average maternal recombination rate is about 1.7x higher than the paternal rate. The full-sibling cluster is almost completely separated from the other relationships, but regardless it’s easy to distinguish them based on fully-identical regions (FIRs).
The Orogen predictor was already giving great predictions for grandparent/grandchild relationships as well as paternal half-siblings because those relationships can have very high or very low cMs. So in a way it isn’t surprising that SegcM now does amazingly well for those relationships. Adding in the segments provides enough distinction between clusters that maternal grandparent/grandchild or half-sibling sometimes have over 50% probability, paternal half-siblings can have much more, and values of paternal grandparent/grandchild have already been found to have 100% probability.
The avuncular relationships (both maternal and paternal aunt/uncle/niece/nephew) have much more overlap with each other and with other relationships. For all of the visible overlap in the plot, you can see that the number of segments splits the group into three possibilities for a large chunk of the top part of the graph and a different trio of possibilities for most of the bottom part of the graph. CMs, which we already knew are more important, do the rest of the work.
Methodology
You might be wondering how I was able to develop relationship predictions from two different independent variables, total cMs and number of segments. There’ve been regression and classification algorithms around for several decades and that’s what we called them: regression and classification algorithms. But I guess we’re calling them machine learning models now. The goal in this case is to separate the different relationship types, or classes, using the best fitting hyperplane.
Preparing data, choosing regression parameters, and compiling the output probabilities was a tremendous amount of work, enough to have a noticeable effect on my eyesight, resulting in reading glasses for yours truly. But now that I have all of the code written, saved, and organized, all of the code can be re-run pretty quickly. Python does the really complicated parts automatically. Apart from keeping track of the number of segments, the pre-processing was the same as in Nicholson (2022).
Currently, SegcM only includes predictions for the 3rd cousin group and closer. The benefit of including the number of segments rapidly decreases with lower cMs and it becomes much more difficult for a regression algorithm to separate a dozen groups from each other when they’re all possible over a given range of cMs and segments.
Also, number of segment predictions are only currently available for half-identical region (HIR) sites, which include all except 23andMe. This means that you can get cM and number of segment predictions for any relationships at 23andMe except full-siblings. But you’d have to subtract the X-DNA, just like one would have to for relationship predictions at any site other than DNA-Sci. Sometimes matches at 23andMe aren’t sharing segment information, so we don’t have access to the amount of X-DNA. In that case, you could subtract off the average X-DNA for a given relationship type if you know it’s possible for you to share X-DNA with a particular match. Also, I’m sure I’ll have all of the work done for 23andMe predictions based on number of segments eventually.
The data used for the Orogen predictions came from Ped-sim. In this case, the refined genetic map of Bhérer et al. (2017) was used as well as the crossover interference parameters of Campbell et al. (2015). I compiled 500,000 data points for each relationship type.
Going Forward
For a couple of weeks I’ve had the opportunity to see how this tool will be used to help people solve cases of unknown matches. Clearly this will help adoptees find their close family members more quickly. I’ve seen several cases in which people were looking for close family members and this tool could have provided a clear and quick way forward.
Here are the first five combinations of total cMs and number of segments that I came across from people looking for help once I started beta testing, in the order in which I saw them over just a three day period (i.e. no cherry picking):


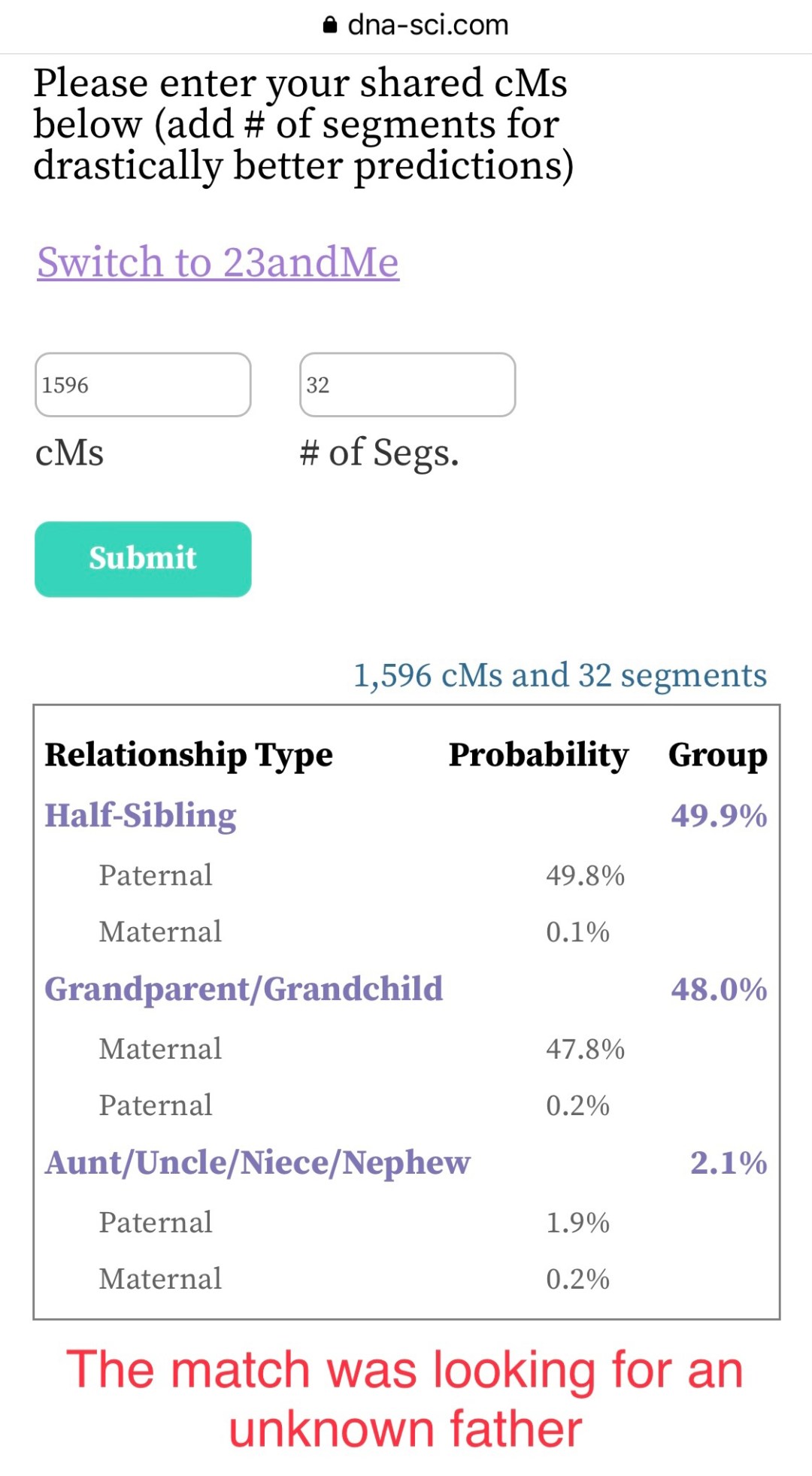


The first person was looking for her father and soon after confirmed that this was a paternal match. There was almost no chance that they were anything other than a paternal half-sibling. As I’ve seen from over 100 empirical data points, the predictions are very accurate. It’s helpful to see the true relationship listed right near the top with a very high probability.
The second person had a match who was much older than her and was on her maternal side. While age isn’t always a good predictor of relationships, this match seemed to be a maternal paternal half- or great-aunt/uncle. Ancestry had them listed as a 1st to 2nd cousin. It’s much more helpful to see the likely relationships listed at the top.
The third person suspected that their father had another child and that this was that person, who was also looking for her father. SegcM suggests that they were right!
The fourth person had a question about a known relative. I asked if they were a maternal maternal 1st cousin and the answer was “yes.”
The fifth person found a match on her father’s side she suspected was a half-sister. They were born two months apart. While it’s always good to confirm that one isn’t the aunt of the other by making sure neither shows as “both sides,” and while this kind of thing definitely happens, it’s less likely that both grandparents or a full-sibling hid their child who was born two months after her. Surprise half-aunts are a bit more common. And does she even have a full-sibling who was old enough to have a child at that time? The predictor suggests that this is her paternal half-sister. Does that sound fairly reasonable?
When I was testing the predictor against known relationships that were sent to me, there was one value for a paternal grandfather with a 0% probability. That concerned me. But I found out a few days later that the relationship was mislabeled. It turned out that the grandfather was actually maternal with a 48.9% probability. So that’s one more benefit of this tool. It might tell you if you need to double-check your notes!
Also, in order for empirical data to help me validate the tool, I needed to know the exact relationship. There were probably a dozen times when I asked the people sending me data “This is a maternal relationship, right?” (or I asked if it was paternal) and they affirmed. There were no cases when I guessed wrong.
Discussion
Previously, the most accurate predictions we could get were from Orogen. That tool predicted the right relationship subtype just over 26% of the time. Using segments along with cMs nearly doubles that, to over 45%. It should be noted that these averages apply to the particular dataset of 125 close matches. If a different dataset included distant cousins or even more first cousins than this one, the averages would be much lower. Similarly, if a dataset included more full-siblings, the averages would be higher. But we can definitely learn by comparing different tools to the same dataset.
We’ve seen how the tools perform when it comes to relationship subtypes. It took me a long time to look into this, but I’ve also recently compared the ability of a few different tools to predict groups. It’s much easier for a tool to say that a relationship is likely in Group 2 (half-sibling, avuncular, etc.).
The functionality of cM-only predictions at SegcM and Orogen is similar to that of a tool at an excellent site called DNA Painter, which uses data that came from simulations conducted by AncestryDNA. The simulations resulted in probabilities that were shown in a graph in their 2016 matching white paper. These probabilities were then copied using an online plot digitizer. Data from Ancestry simulations (DNA Painter) give the correct relationship group an average probability of 97% for the 125 close matches. Orogen predictions gave the true group a probability of 98%. The new predictions at SegcM give the right group an average probability of 99%.
It’s no surprise that relationship predictors get the right group for close family matches. But we’ve seen above many examples of how we can get a greater than 50% probability for the right relationship subtype. It’s great to be living in the future. We’re now thinking about relationship predictions in 2D!
DNA-Sci is also the home of the double cousin predictor and DNA coverage calculator.
DNA-Sci — advancing the science of relationship predictions. Please also submit data to this new DNA match survey that will greatly help improve and build new relationship prediction tools. You can also find mobile apps. for relationship predictions in the Apple Store and on Google Play. Feel free to ask a question or leave a comment. You might also like this tool to visualize how much DNA full-siblings share. DNA-Sci is also the original home of DNA coverage calculations.

That looks like a great tool. If I have values for 3 siblings is it best to take the average value & number of segments? Could the tool be refined further to give in several different sets of sibling values?
Hi Paul,
Thanks for you comment. And this is something I’ve been wanting to write about for the past couple of days. I’ve wanted to add some functionality for this for a long time now. I’ve actually done a whole study on the art of using multiple siblings’ kits, this is the third of three parts: https://dna-sci.com/2022/01/10/your-siblings-have-dna-kits-too-part-3-empirical-data/
When you don’t have chromosome browser information, the best thing to do is to take the average of the kits. When you can, the best thing to do (except for one other method that’s pretty difficult) is add up the distinct segments using a tool from Jonny Perl and then multiply by a ratio, such as 100%/75% for two kits, 100%/87.5% for three kits, etc.
But there are some catches even to the tried and true methods. Let’s say you have an unknown match that appears to be a paternal half-sibling because they share a very large amount of DNA in the Group 2 range. And then your sibling matches the person on the low end of the Group 2 range, which also could suggest paternal half-siblings with high likelihood. Well, if you average the two, you’ll get a middle of the road Group 2 cM value that will probably be more suggestive of a maternal half-sibling or avuncular relationship.
Adding in the number of segments gives us a much more refined picture of the possible relationships. We don’t want to ruin that by averaging. It may be that, while “twinning” went from the preferred method, to a method proved to be the worst performing of all, that it will come back to be the best method when breaking close family relationships down by subtypes and using the number of segments for predictions.
My instincts tell me that, for more distant matches, we could average both the cMs and the segments. But I would have to do some testing to be sure.
I was successfully using the centimorgan plus number of segments tool until yesterday when using the tool started requiring a password. Am I supposed to be entering a password or is this a glitch? Thank you! I love this tool.
Hi Susan,
I’m really sorry. I had to password protect the prediction tools because there’s a very poorly-designed comparison of relationship predictors being done right now. The comparison incentivizes predictors lumping more relationships into groups. It looks at the probability assigned to the correct group and it records whether or not a predictor shows the correct relationship in the top three probabilities.
So one can think about it this way: if I built a tool today that said “100% for all relationship types” no matter what the user input, this predictor would look perfect in the comparison. It would always get a score of 100%. And it would always get the right relationship within the top three possibilities (actually the top one possibility).
The results of this comparison will look very favorably on the 2016 Ancestry simulated probabilities because they lump more relationships into bigger groups than any other predictor.
I don’t know if this comparison is designed intentionally or not to be biased against the SegcM and MyHeritage probabilities. The two authors have their names on a tool that shows the Ancestry probabilities. And they’ve both shown bias against DNA-Sci tools in the past. One of the authors also has a personal issue with the designer of the MyHeritage tool.
But whether or not the bias is intentional, it’s still there. And the community needs to speak out against it. It’s really unfair that adoptees might not be getting their answers right now and other people might not know how their close family matches might be related to them.
I hope the tools can come back fully online soon.
Also, I’m going to send you an email.
This is very unfortunate, is there someone we can contact to register a complaint about this comparison?
Hello,
Yesterday there was a good development in that one of the designers of the comparison said he’s received “many” complaints about the biases in the design and is “proceeding accordingly.” That language isn’t strong enough for me, but it’s a step in the right direction. There was also a statement that this is how science is conducted. This is, in fact, not how science is conducted. Scientists don’t publicize their intent to proceed with an extremely biased comparison, refuse criticism about it from the people who are collaborating with them for several days, and then continue to be vague about how the methodology will be changed. I think that people should keep voicing their concerns.
Also, the other designer of the comparison has recently expressed that she has no desire to account for the differences between individual relationships and relationship groups, saying that if a tool reports lower probabilities for individual relationships than another tool reports for group probabilities, then the first tool should be penalized for the lower probabilities. This betrays a fundamental misunderstanding of probabilities.
Another good development is that yesterday the page for SegcM in the comparison survey changed to say “ACCESS TO THIS TOOL HAS BEEN REMOVED BY THE DEVELOPER, SO IT CAN NO LONGER BE EVALUATED” whereas it used to tell respondents to keep trying the link. The password protection has been removed as of today, so you can now access the SegcM tool as much as you’d like.
The more we raise awareness about these issues, the better. People should understand what this study was proposing. It’s impossible for the individual relationship probabilities from one tool to be greater than the probabilities of a group of relationships at another tool. It’s also impossible for a tool that shows individual relationship probabilities to show the correct relationship within the top three possibilities more often than a tool that lumps more relationship types into groups. Whether this was intentional or because of a very poor understanding of probabilities, this is very concerning. Genetic genealogists shouldn’t let their guards down about study was set up this way until the problem has been completely remedied.
Even if SegcM is going to be left out of this comparison, MyHeritage will likely be maligned, even more so than SegcM would have been. Data for the metric of “how often is the correct relationship found within the top three probabilities” have already been collected. The only proper solution is for all of those data to be thrown out, but it’s hard to believe that the study authors would do that.
Genetic genealogists should speak out against this unfair treatment of MyHeritage both before and after this study is released. The more they do, the less chance that that metric will be published and the less credibility the study results will be given after they’re released. Without a statement from the comparison designers indicating that they don’t intend to use this metric, we have every reason to believe that they plan to proceed.
May I also have a password?
Hi Leona,
The password protection was removed today.
Can I have the password? Thank you.
Hello,
The password protection was removed today.
I have been using SegcM for the last couple of weeks to add notes to Ancestry matches that are fairly close matches but have no trees and whom I can’t place otherwise. This is so that at least I have a rough idea where they fit. As part of this process I have been post-filtering the groups to fit my actual tree (for example my grandparents all died long before dna testing so I know it can’t be them). I realise this might also eliminate NPE surprises but I find it useful anyway.
It occurs to me that any elimination of possible relationships would affect the probabilities, as indeed would the actual numbers of each relationship (for example if I have only one 1st cousin but 40 2nd cousins this would alter the probabilities from the norm)
Am I right in this thinking, and if so do you think it is possible that some future version of this program could be fed a Gedcom file and produce either a pre-filtered or pre-weighted version of the result tailored to individual family trees ?
Hi Steve,
That kind of functionality should be possible. But I have to wonder, if someone knows that much about their match, they probably already know exactly how they fit into their tree and have no need to use a relationship predictor.
Probabilities at DNA-Sci have already been using population weights for a couple of years. You can see in this article that a person might have, on average, 2.5 siblings, 3 aunts/uncles, 7.5 1st cousins, 37.5 2nd cousins, etc.: https://www.sciencedirect.com/science/article/pii/S1875176822000099?fbclid=IwAR3FNCK21PVh9Nnq0T7U3LfaVBRdEgsXDP9A8fMevAtnvu4SYISXGGUsuyw
That’s why DNA-Sci predictors, 2016 Ancestry simulated probabilities say that the most probable relationship is something like a 6th to 9th cousin for 8 cMs. Probabilities that don’t use population weights like MyHeritage and newer (2019 to present) Ancestry probabilities show something more like a 4th cousin.
When it says mat.pat, mat.pat x 2
Does that mean for first cousins that their parents would have to be brother and sister.
Apologies if the question sounds dumb, just want to double check and make sure I’m understanding correctly. Relationship I’m researching is 1277cm 40seg
Hi Tara,
Yes, your maternal paternal 1st cousin is your mother’s brother’s child. Your paternal maternal 1st cousin is your father’s sister’s child. 1st cousin is the most likely relationship for your shared DNA. There’s a 9.6% probability that they’re your mother’s brother’s child and an additional 9.6% probability that they’re your father’s sister’s child. The x2 is just there to remind people that if they only add that percentage one time they won’t get 100% for the total of all probabilities.
It works a little differently for some other relationships. For example a maternal paternal great aunt is your mother’s father’s sibling. So in this case the gender path stays vertical in your tree the entire time instead of jumping over to a sibling like for 1st cousins. The sex of the testers doesn’t matter and we don’t need to worry about ancestor pairs. So it just starts at you and follows any applicable genders that you have to go through to get to your match.
Hi there, I have two relatives in my ancestry. We share 5x great-grandparents on my maternal side.
One of the relatives is a male we share less than 1% DNA: 12cM across 2 segments
The other is a female, we share less than 1% DNA: 54 cM across 4 segments. In the simplest terms, what does this mean?
Hi Lynn,
A segment can be a whole chromosome or any smaller part of a chromosome. You can also share several segments with a person on one chromosome. Or if you share four segments with someone there’s a good chance that they could all be on different chromosomes. When you share 12 cMs with someone on two segments that means you share something like two 6 cM segments. That wouldn’t happen at a place like Ancestry because they cut off segments less than 8 cMs unless you saved your match before they changed it to 8 cMs. There are multiple ways you could share 54 cMs over four segments with someone. For example, you could share four segment that are each about 13 cMs. Or you could share one 8 cM segment, one 12 cM segment, one 15 cM segment, and one 19 cM segment. There are infinite possibilities.
Hello! Thanks for this brilliant tool! I wondered if you could help me please?
I do not know who my biological father is or any of his family as I was an AI baby. I have found some very close matches on Ancestry and have run them through the tool, which is extremely helpful. I’ve found that one match is either:
Grandparent/Grandchild, Maternal 49.5%
Half-Sibling, Paternal 49.4%
Given that I know this is my biological father’s line does that mean the match must be a half-sibling?
The match is listed n Ancestry as being female but it’s possible when I cropped up things were changed to protect identities given a family tree was unlinked very swiftly (and unfortunately I didn’t look at it quickly enough!) So I’m trying not to assume anything and go with the data.
Many thanks for any help you can give me!
Hello,
Yes, that’s almost certainly a paternal half sibling. There might be something like a 1/1000 chance of aunt/niece. Do they respond to messages? It’d be good for you to both check for “paternal” labels. If either of you has a “both sides” label, that might indicate a different relationship. If you don’t have labels, you could both do the Leeds method. Also, if you’re female, it’s a good idea to upload to GEDmatch and check the X Chromosome. Paternal half sisters will always share a full copy of X-DNA.